Upon Matrix and Graph Representations of Factor Graphs
Motivation
- Understand better the connections between matrices and graphs in previous works; Recap details.
- Factor Graphs for Robot Perception
- Have a glance at advances in the intersection of graph theory and SLAM
- Reliable Graphs for SLAM, IJRR 2019
- Cramér–Rao Bounds and Optimal Design Metrics for Pose-Graph SLAM, TRO 2021
Factor Graph and its Information Matrix

We start with revisiting a factor graph and its corresponding Jacobian
- Each factor corresponds to several rows. It is a measurement.
- Each variable corresponds to several columns. It is a state to be estimated.
So we have a factor graph, and its corresponding linear system \(Ax = b\).
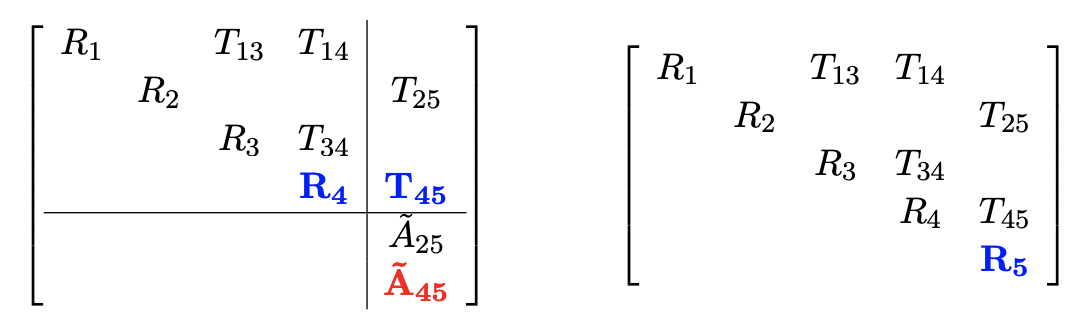
Variable elimination and matrix factorization
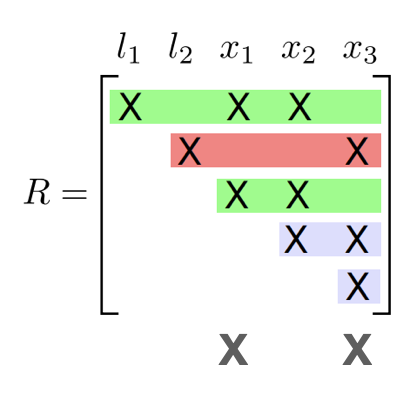
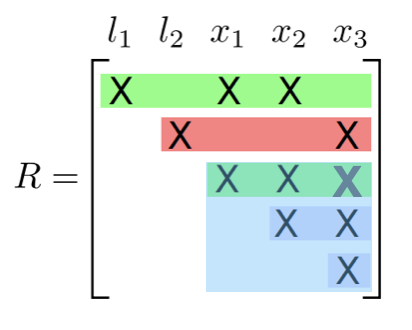
In large sparse SLAM setups, we usually factorize A with QR. Partial QR is iteratively applied to submatrices, with permutation (?).
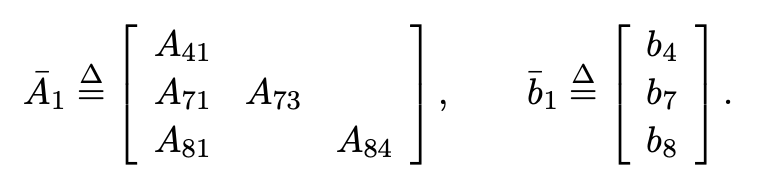
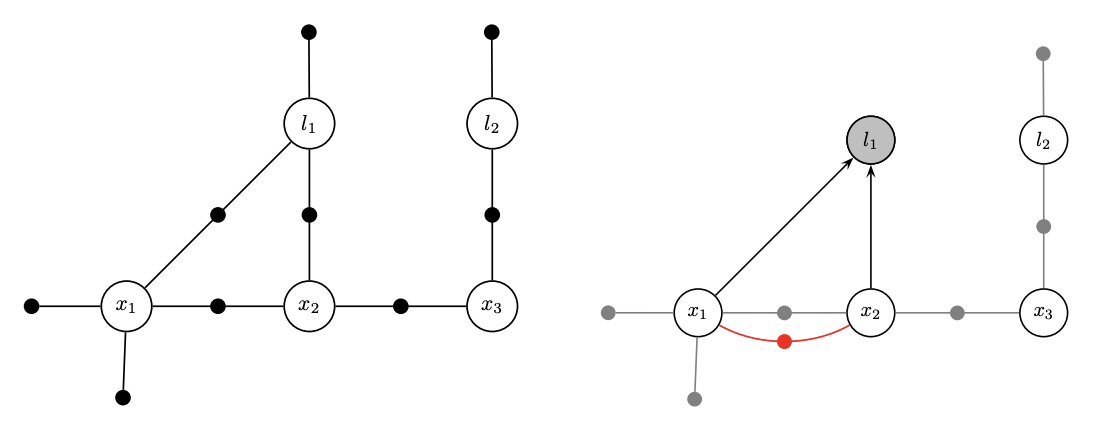
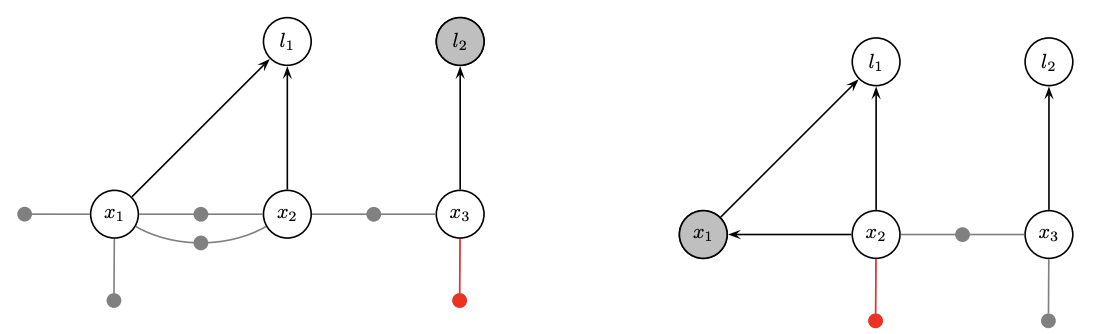
- Factorization is variable elimination in a graph
- The spirit is like ‘an undirected graph to a DAG’
- Factorization is partial QR in a matrix
- Use Householder reflections or Givens rotations
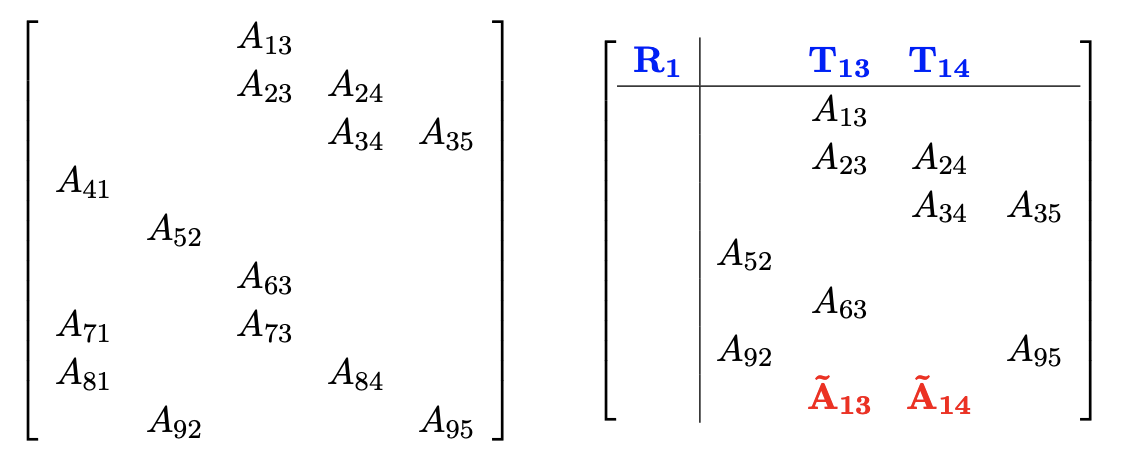
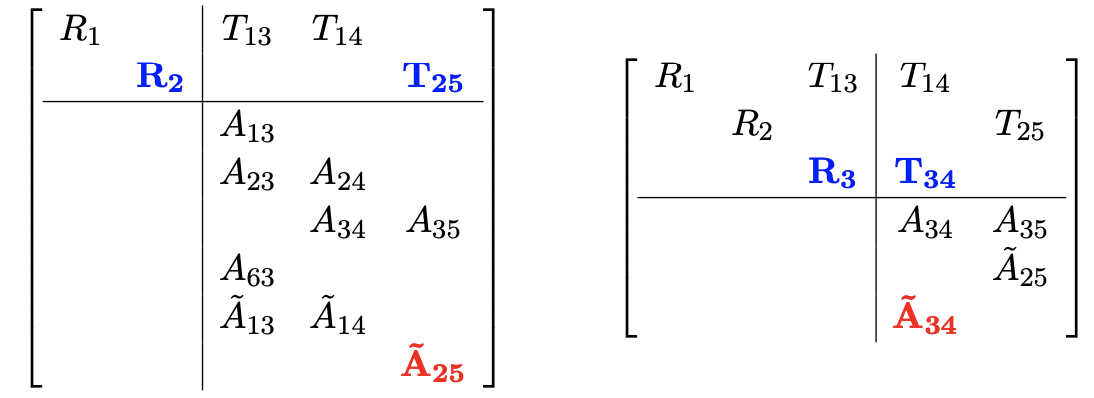
Variable elimination ordering
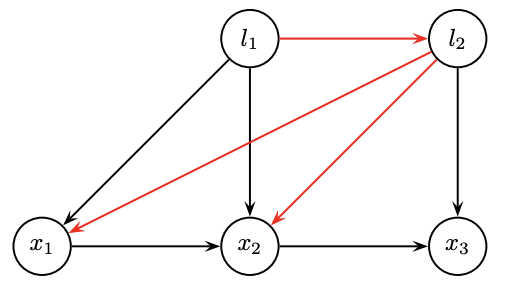
The ordering in elimination matters. A bad order results in fill-in and leads to a dense matrix to solve.
- Topologically densely connected
-
We prefer smaller degrees in order

- Degrees:
- l1=1, l2=1; x1=2; x2=3, x3=2.
-
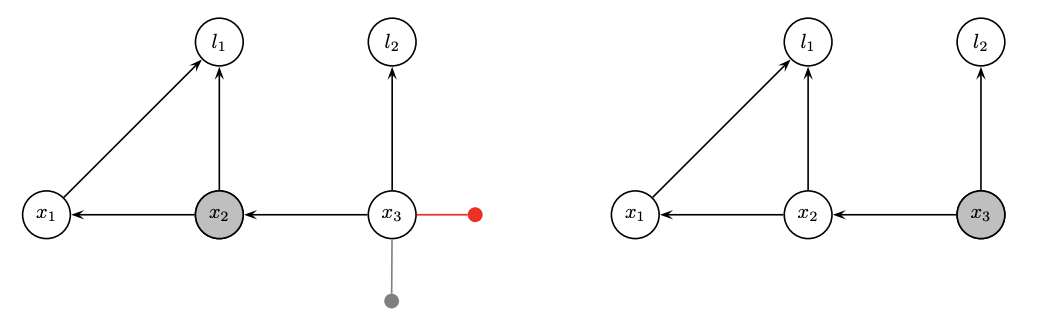
Computationally dense
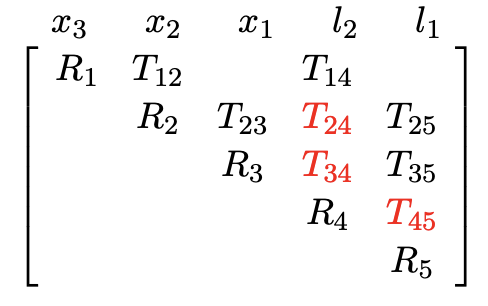
- We prefer fewer entries in the rows during QR
- Direct order: 2 fill-in, then 1 fill-in
- Inverse order: 2 fill-in, then 3 fill-in
Heuristics for reordering
-
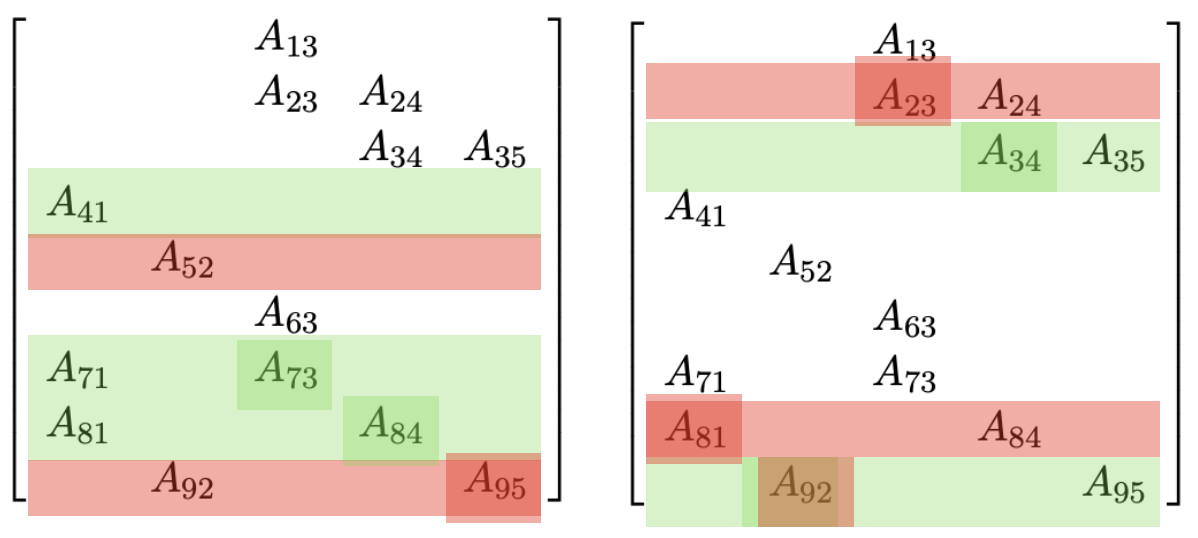
Reorder according to degrees: COLAMD
[TODO] https://citeseerx.ist.psu.edu/viewdoc/download?doi=10.1.1.124.1109&rep=rep1&type=pdf
- Graph partitioning: METIS
-
A graph can be somehow partitioned into
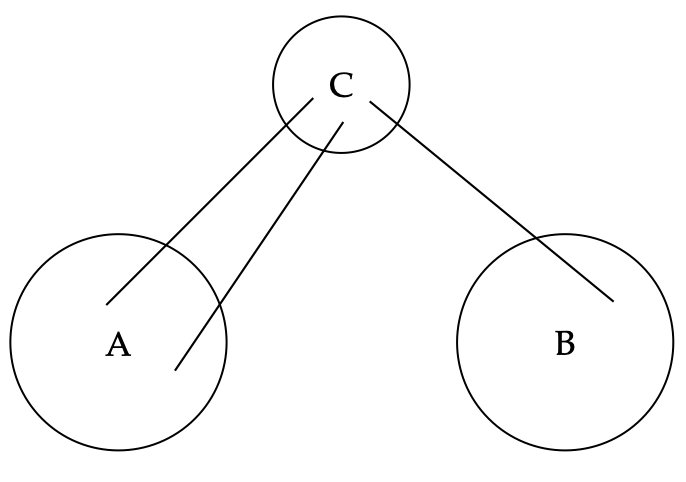
Incremental update and the clique tree
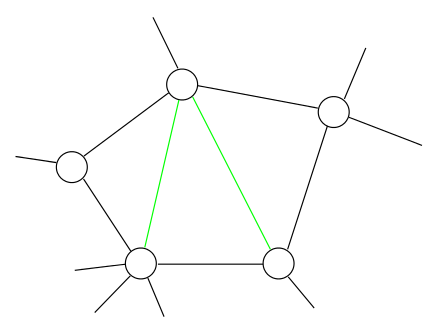
Fact 1
- Variable elimination results in not only a DAG but also a chordal graph (PGM Theorem 9.8)
- A chordal graph is a triangulated graph
Fact 2
-
We can form a clique graph from a graph, by connecting max-cliques
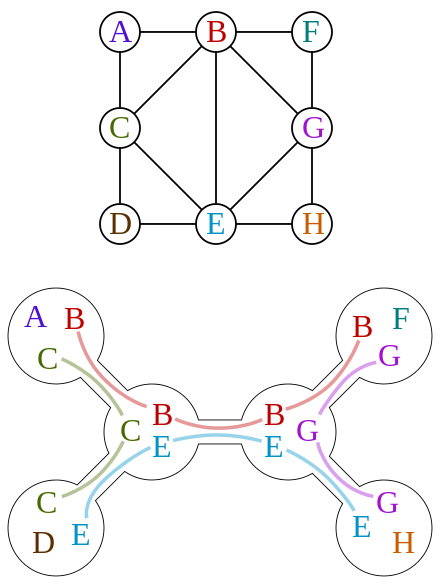
Fact 3
- A clique graph is a clique tree in a chordal graph
-
Trees are good for elimination or factorization esp. when the cliques are small
-
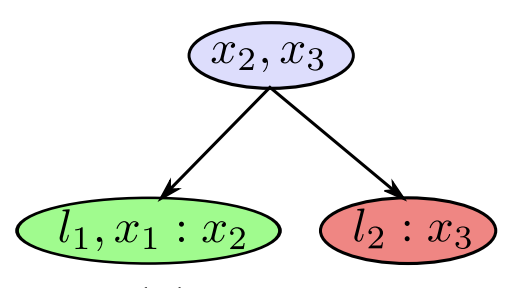
A clique tree depicts the global structure
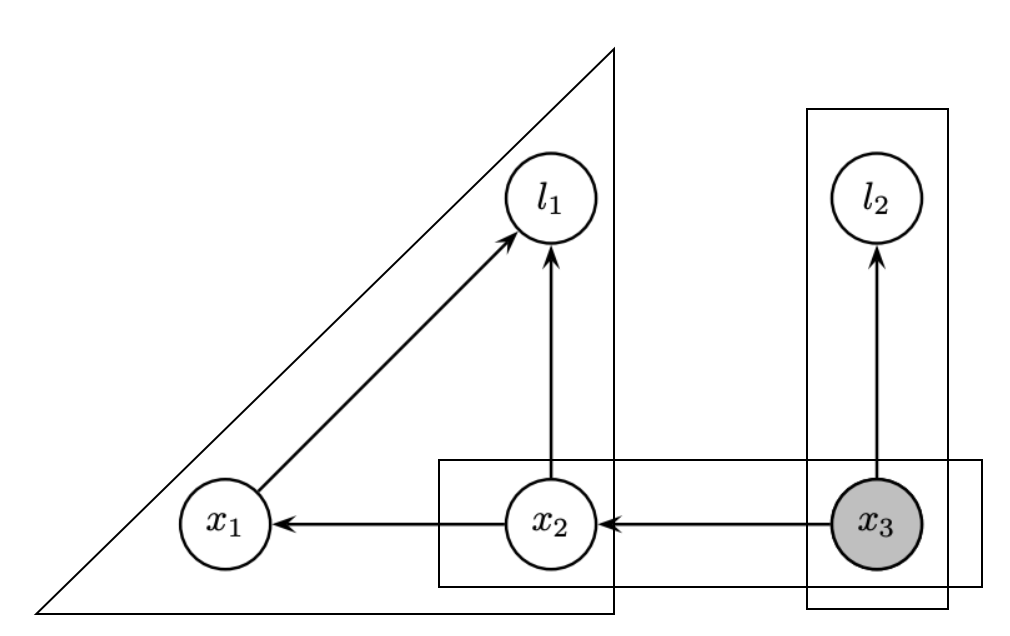
-
Each clique depicts local distributions
-
In a clique is a dense upper triangular matrix after reducing the separator column
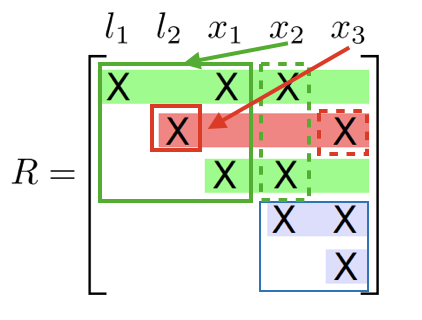
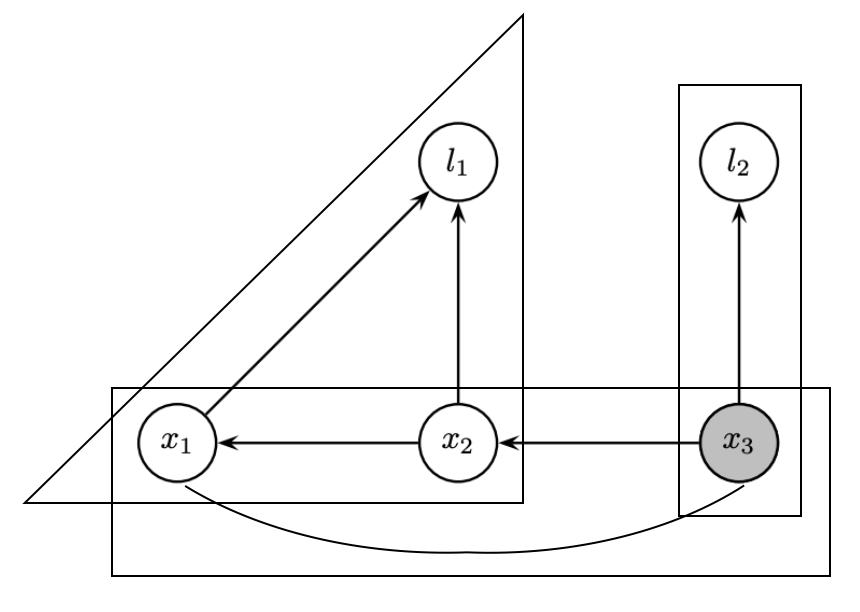
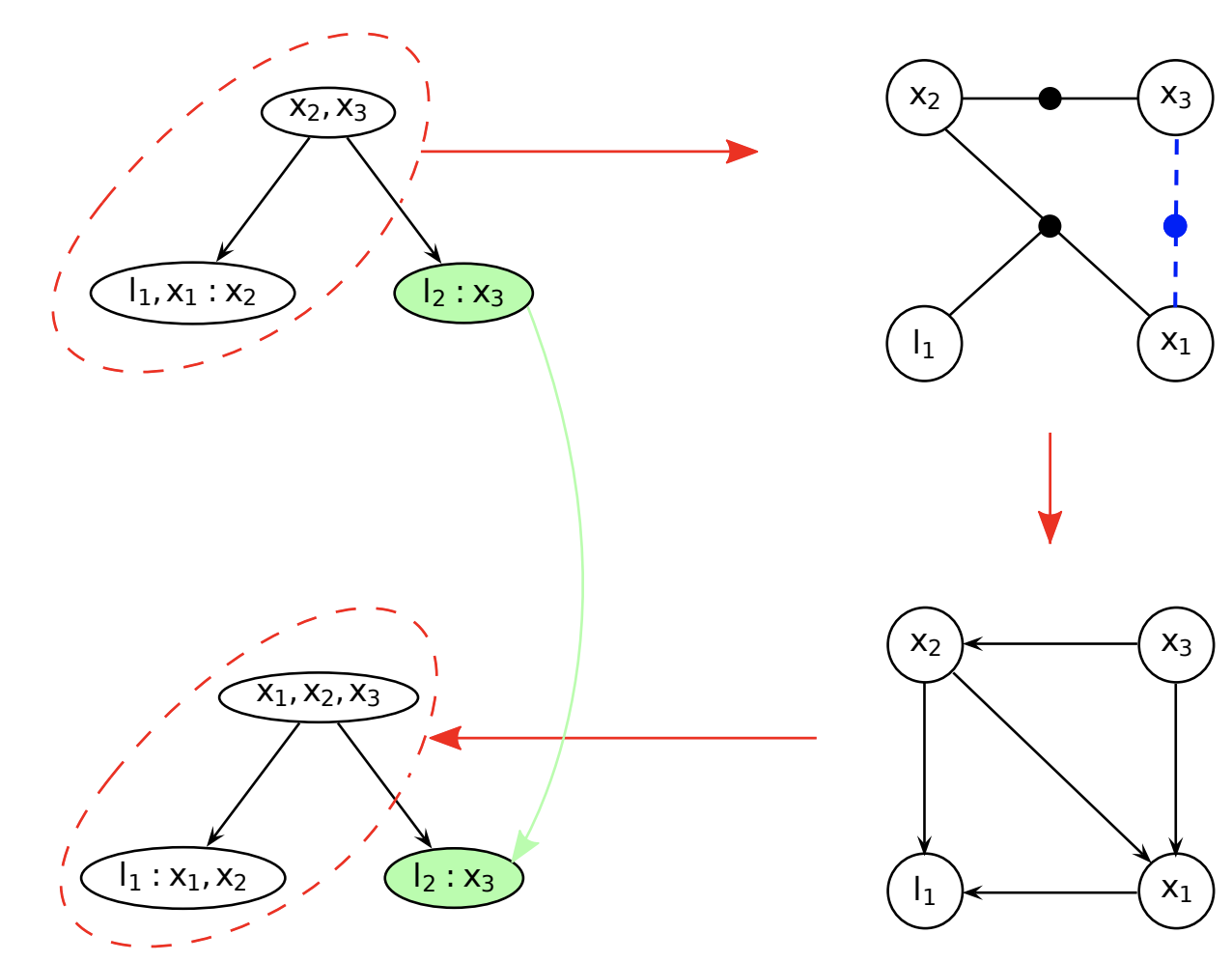
Incremental update
- Identify the path from the root that contains the variables
- Redo the variable elimination locally to update the clique structure
- Givens rotation locally
Summary
Up to now, we know the connection between
- Matrix factorization (QR decomposition, reordering rows, locally dense triangles)
- and graph topology manipulation (undirected to DAG, factor to Bayes, degree, clique)
Heuristically, we know local topology (vertex degree) matters. There could be more fundamental insights.
Depict a graph in matrix
- We used graphics to depict graphs. They are intuitive, but not easy to analyze.
- Although they are related to Jacobian matrices (in factorization), they are not equivalent.
- We now use incidence matrices and graph laplacian to directly depict matrix topology.
Incidence matrix
-
A V x E matrix, each column encode the (weighted) edge connecting 2 vertices
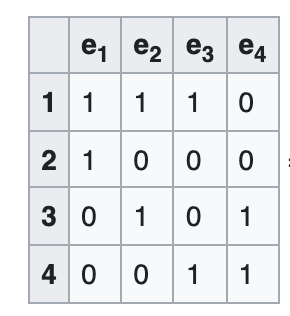
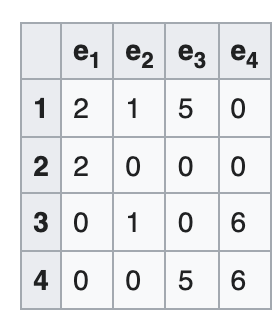
-
Sounds familiar? A Jacobian is E x V, each row encodes a measurement connecting 2 variables


There are even more similarities.
Laplacian matrix
- Laplacian is given by \(L = D - A\), where D and A are degree and adjacency matrices. It is a V x V matrix.
- Graph laplacian depicts the graph’s intrinsic connectivity at the vertices’ perspective.
-
Another derivation is \(L = MM^\top\), where \(M\) is the incidence matrix.
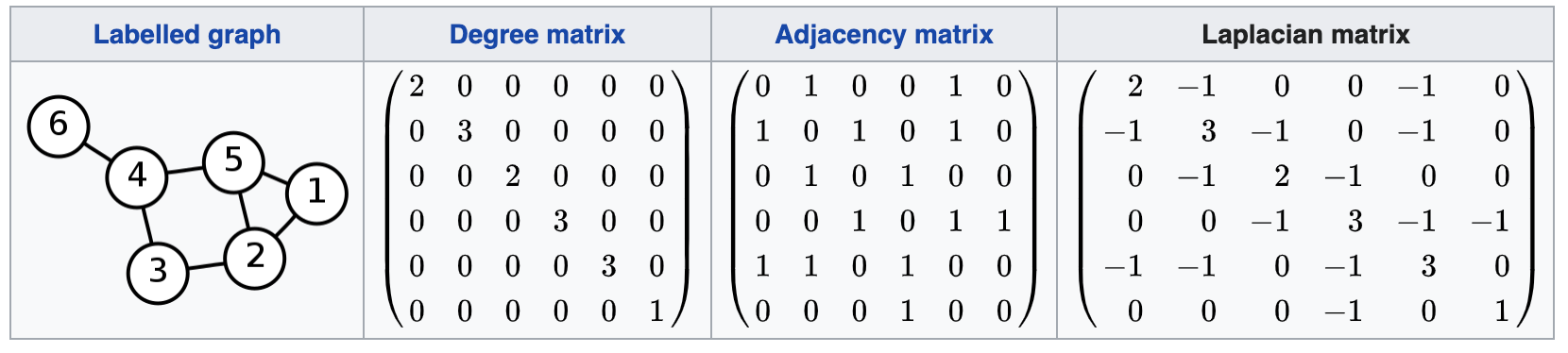
- Sounds familiar again? A Fisher Information Matrix is given by \(\Lambda = J^\top J\), where \(J\) is the Jacobian matrix.
- We will revisit some special setups where \(\Lambda\) and \(L\) can be directly connected with equations.
Conectivity from Laplacian
- Key message: we can obtain global topology (graph **connectivity) of the graph via the Laplacian matrix.
- Measurement 1: Algebraic connectivity / Fielder value
- \(\lambda_2(L)\), where \(0 = \lambda_1 \le \cdots \le \lambda_n\)
- This connecivity is 0 if the graph is disconnected; 1 if the graph is complete.
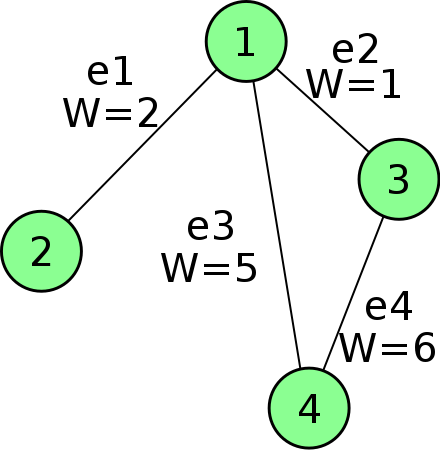
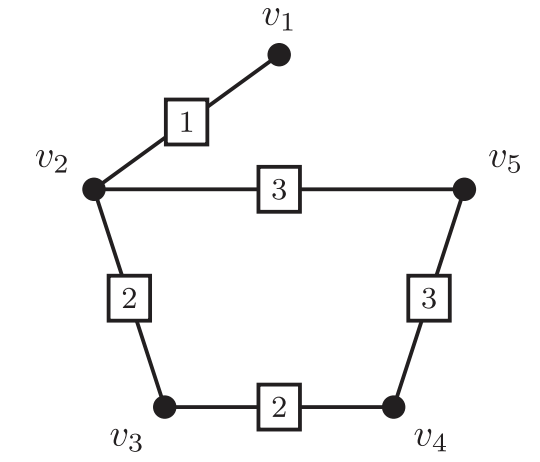
- For the example above the eigen values are:
- [3.33066907e-16, 7.21586391e-01, 1.68256939e+00, 3.00000000e+00, 3.70462437e+00, 4.89121985e+00]
- Associated eigenvector:
- [0.41486979, 0.30944167, 0.0692328 , -0.22093352, 0.22093352, -0.79354426]
- Measurement 2: Tree connectivity / Kirchoff’s matrix-tree theorem
- Count the number of spanning trees
- \[t(G) = \frac{1}{n} \lambda_2 \lambda_3 \cdots\lambda_n\]
- For the example above: 11
- Proof: http://math.fau.edu/locke/Graphmat.htm
- Related: Cayley’s formula \(t(G) = n^{n-2}\) for a complete graph.
- Count the number of spanning trees
Reduced Laplacian
- Remove one selected row (vertex) and the corresponding arbitrary column (edge)
- ‘Corresponding to’ a Jacobian: remove a vertex or anchor a variable
- Algebraic connectivity: \(\lambda_1(L^r)\)
- SVD / Eigenvalue decomposition
- Tree connectivity: \(\det L^r\)
- Relatively easy to compute, esp. for sparse matrices:
- Use Cholesky decomposition, then compute the product of diagonal elements
- Relatively easy to compute, esp. for sparse matrices:
Associate Laplacian with Information
- The simplest \(R^d\)-sync problem
- We have variables \(x_j \in \mathbb{R}^d\) and measurements \(z_{ij} = x_i - x_j + \epsilon_{ij}\).
- Covariance per edge is simplified to \(\sigma_{i}^2 \mathbf{I}_d\)
- The Jacobian is the incidence matrix (transpose), if d=1 and the variance per edge is 1 and we discard priors

-
If we take in one prior then the Jacobian is the reduced incident matrix (transpose)
\[z = \mathbf{M}^{r, \top} x + \epsilon\] -
If we extend to arbitrary dimension d then it becomes
\(z = (\mathbf{M}^r \otimes \mathbf{I}_d)^\top x + \epsilon\) (Kronecker multipler expands each 1x1 entry to a dxd block)
-
Then considering the variance as a diagonal weight matrix, the information matrix is given by
\[\Lambda = (\mathbf{M}^r \otimes \mathbf{I}_d)^\top (\mathbf{W} \otimes \mathbf{I}_d) (\mathbf{M}^r \otimes \mathbf{I}_d) = (\mathbf{M}^r \mathbf{W} \mathbf{M}^r)^\top \otimes \mathbf{I}_d = \mathbf{L}^r_w \otimes \mathbf{I}_d\]Here we are using the weighted Laplacian, where the weight per edge is given by the measurement variance.
-
This is a very close association between Information and Laplacian matrices.
\[\Lambda = \mathbf{L}^r_w \otimes \mathbf{I}_d\]
E-Optimal and Albegraic connectivity
- Covariance is inverse information \(\mathbf{C}^{-1} \approx \Lambda = \mathbf{L}^r_w \otimes \mathbf{I}_d\)
- If we apply eigenvalue decomposition to a covariance matrix C, then the max eigenvalue gives the worst case error variance (E-optimal design) among all unit directions.
- \[\lambda_{max}(C) \approx 1 / \lambda_1(\Lambda) = 1 / \lambda_1(L_w^r)\]
-
So the albebraic connectivity defines the variance error bound
D-criterion and Tree connectivity
- If we apply determinant to a covariance matrix C, then the value provides a scalar measure of the uncertainty (D-criterion) in the covariance matrix.
- \[\log \det C \approx \log \det \Lambda^{-1} = - \log \det (\mathbf{L}^r_w \otimes \mathbf{I}_d) = -d \log t(G)\]
- So the tree connectivity gives the uncertainty estimate
There are extensions to less trivial 2D/3D SLAM, and corresponding corollaries. In a nutshell, graph topology and estimation uncertainty is connected via Laplacian and Fisher Information.
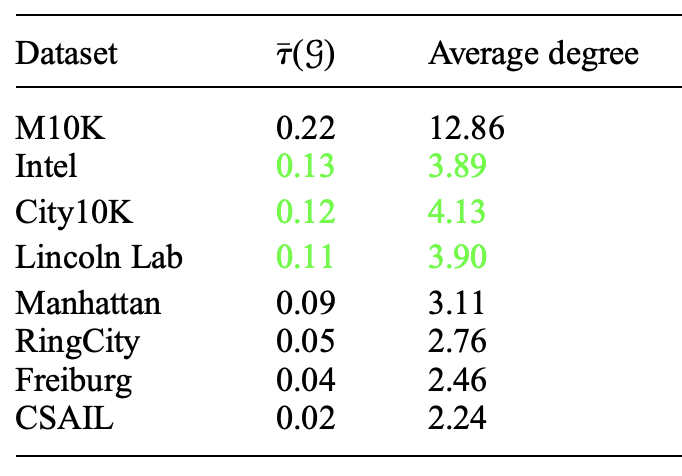
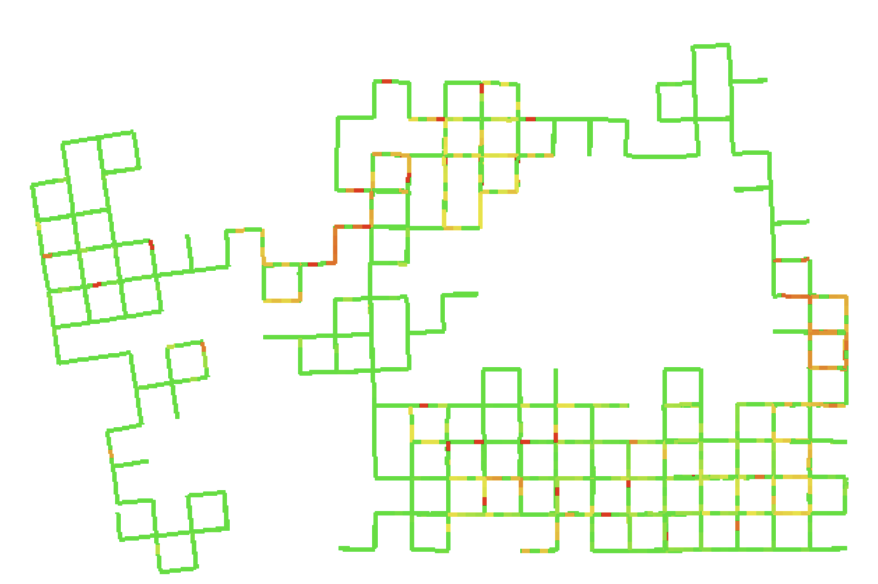
Topology selection for a better D-criterion
With the empirical connection, we drop covariance now and consider only the topology or the Laplacian
We are interested in edge selection (e.g., loop closure selection in a pose graph)
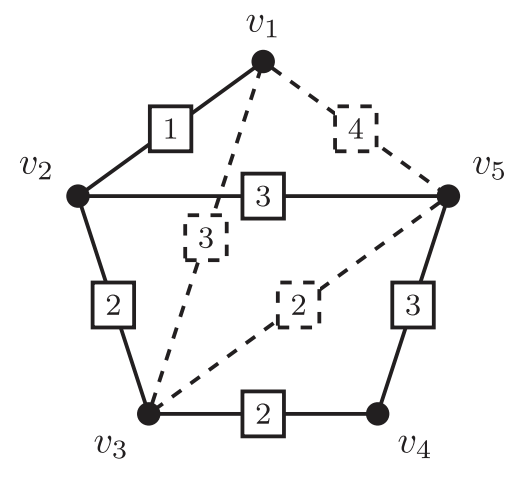
- k-ESP: Given a initial graph and several candidate edges (say potential loop closures), select at most k-edges to maximize connectivity and reduce uncertainty
- \(\Delta\)-ESP: Select as few edges as possible to reach the desired information gain \(\Delta\)
We know this can be computed in a batch:
- Select several edges, run Cholesky of the updated Laplacian, and measure t(G) (tree connectivity)
- Exact solution by evaluating in a batches is NP-hard. There are C(m, k) combinations.
Can we do it incrementally by adding one edge $m_e$ in the incidence matrix?
- Yes, the gain is given by \(\Delta_e = m_e^\top L^{-1} m_e\), where \(m_e\) is the additional column in the incidence matrix
-
So we can use the greedy algorithm: every time we add the edge with the max gain, then update the Laplacian
-
Other solutions include convex relaxation
\[L = L_{init} + \sum_j \pi_j L_{e_j} = MW^\pi M\]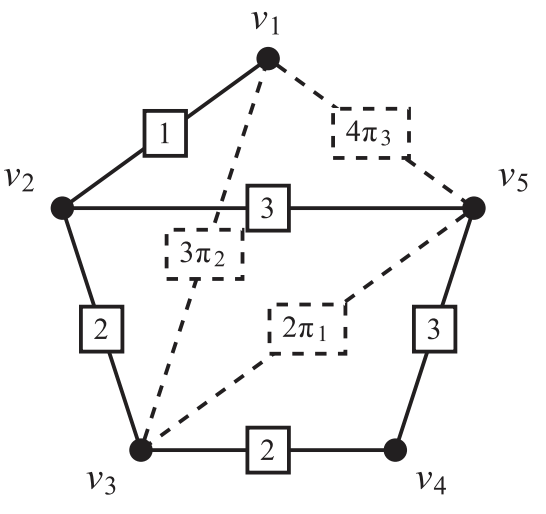
-
Non-convex version
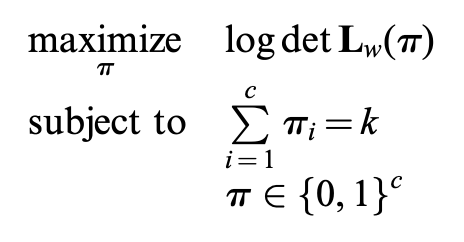
-
Convex relaxation (can be solved with a laplacian multiplier
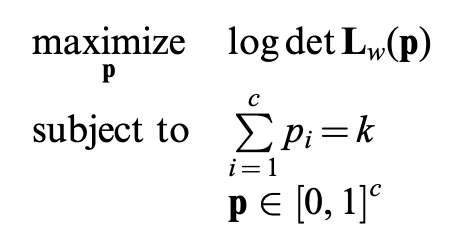
- In short: we know a criteria to properly select edges (loops) by (weighted) topology.
Problems
- Can we generalize from the block diagonal covariance to general covariance? In other words, can we put more information to edges rather than a scalar weight?
- Can we put noisy-or factors on edges to support multi-hypothesis inference?
- Tree-connectivity is a global measurement. Can we limit it in a subgraph and consider properties in clique trees?
- For a clique we can use Cayley’s formula to directly obtain tree connectivity without decomposition. can this help in connectivity measurement?
